
General information for recommended input requirements and instructions for submitting samples, based on sample types.
The Advanced Genomics Core prefers >200 ng for most assays. The higher input yields better performing assays and more robust data.
| Illumina Assay | Min Recommended Input | Min Volume | Quality and Concentration Minimums |
| Poly-A (standard) Library Prep | 50 ng | 25 µl | RIN > 7 and 2 ng/µl |
| Poly-A (low input) Library Prep | 1 ng | 25 µl | RIN > 7 and 0.05 ng/µl |
| Total RNA* (standard) Library Prep | 100-400 ng | 15 µl | DV200 >70% – 6.6 ng/µl (100 ng total) DV200 50-70% – 13.3 ng/µl (200 ng total) DV200 30-50% – 26.7 ng/µl (400 ng total) DV200 < 30% not recommended |
| Total RNA* (low input) Library Prep | 1-4 ng | 15 µl | DV200 >70% – 0.06 ng/µl (100 ng total) DV200 50-70% – 0.13 ng/µl (200 ng total) DV200 30-50% – 0.27 ng/µl (400 ng total) DV200 < 30% not recommended |
| small RNA Library Prep | 10 ng | 15 µl | if submitting total RNA RIN > 7 |
| DNA (standard) Library Prep | 100 ng | 25 µl | 4 ng/µl |
| DNA (low input) Library Prep | 0.5 ng | 25 µl | 0.02 ng/µl |
| User-made Library | 10nM | 25 µl | < 1% adapter dimer |
*The ribosomal depletion probes used during Total RNA library prep are selected based on the species indicated in the submission form. If both bacterial and human/mouse/rat probes are required, you must include both species in your form or indicate that it is mixed species. If your samples are isolated from blood and you require globin depletion, please indicate that in the submission notes.
Preferred Buffers:
- RNA (DNase treated) – molecular biology grade water (NOT DEPC)
- DNA – EB buffer (10mM TRIS (pH= 8.0-8.4)) or molecular biology grade water
- Illumina Libraries – EBT (10mM TRIS + 0.1%Tween20), EB (10mM TRIS)
Note: The presence of EDTA can inhibit enzymatic activity and lead to reduced final yield, failure of library prep, or low and uneven coverage when analyzed bioinformatically.
Oxford Nanopore library preparation is sensitive to contaminants that can be revealed by Nanodrop readings. Note that the rapid chemistry is optimized for samples >30kb, while ligation sequencing can accommodate both HMW DNA and amplicons.
| ONT Assay | Recommended Input | Min Concentration | Min Volume | Quality |
| Ligation Sequencing (with or without Native Barcoding) | 1-2µg | 25 ng/µl | 30-50 µl | 260/280 ~1.8-2.0 260/230 ~2.0-2.2 |
| Rapid Barcoding Sequencing | 500ng (≤12 samples) 100ng (>12 samples) | 55 ng/µl (≤12 samples) 6 ng/µl (>12 samples) | 10-15 µl | 260/280 ~1.8-2.0 260/230 ~2.0-2.2 |
Preferred Buffers:
- 10 mM TRIS (pH=8.0-8.4)
Note: We require that you provide us with a minimum 20ul aliquot of your elution buffer, so that we can use it to get accurate nanodrop readings when assessing the purity of your samples. This aliquot should be labeled with your Service Request ID and “EB”.
| Other Assay | Min Recommended Input | Min Volume |
| DNA/RNA Quality Control | NA | 5 µl* |
| BeadArray Genotyping | 500 ng | 10-15 µl |
| Scroll Quality Assessment | 2-5 scrolls | NA |
* if less than 5 µl is provided, water will be added to your samples to bring the volume to 5 µl
** Depends on sample type and size - For OCT-embedded tissue, we request three - five 10 µm sections. For FFPE, we request two - four 5 µm sections.
Preferred Buffers:
- 10x Genomics – 1X PBS (calcium and magnesium free) containing 0.04% weight/volume BSA (400 μg/ml). If necessary, PBS can be replaced with most common cell culture buffers.
Note: Nanodrop can overestimate the amount of nucleic acid in a sample since it is not DNA/RNA-specific and can be influenced by the presence of contaminants. Fluorescence-based concentrations can be as much as 50% lower than those determined by nanodrop. If using nanodrop to quantify your samples prior to submitting to the AGC, be sure to at least double the amount of material being submitted.
The AGC accepts aliquots in either tubes or plates. You must select either tubes or plates when you create your Service Request in MiCores. You can use either plates or tubes, as you prefer. We recommend using a plate when you have more than 7 aliquots in your submission. When submitting samples in plates, please use 96-well, skirted PCR plates.
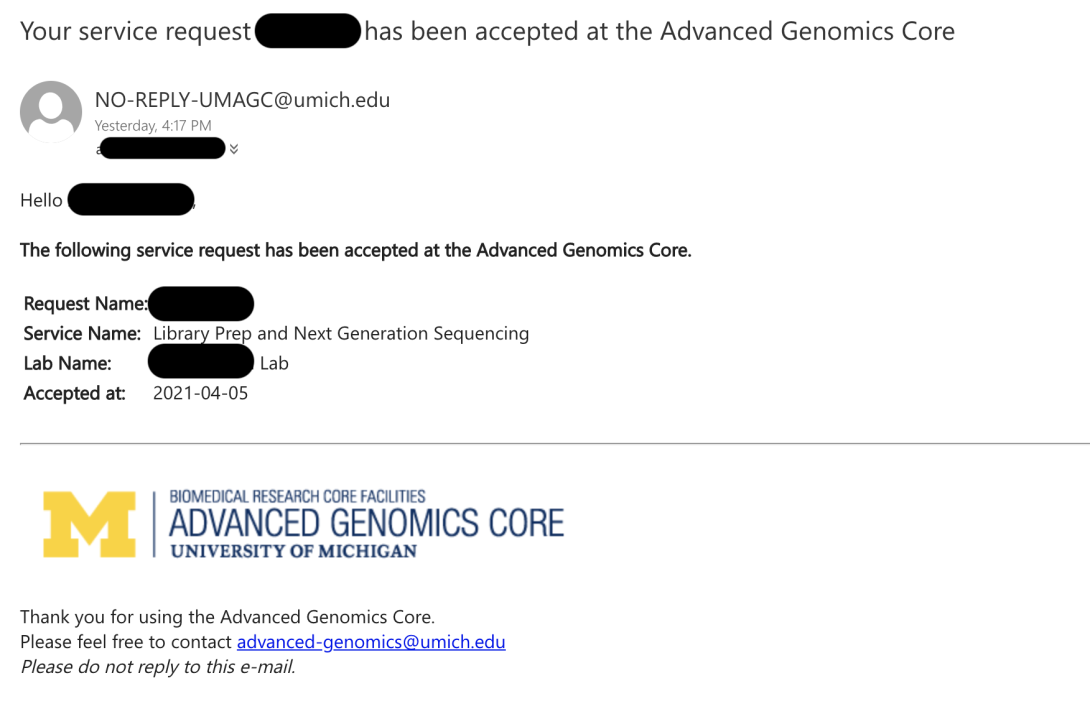
Once your Service Request has been accepted by the core, you will get an email with the Core Sample Ids or Core Plate Ids. You must label your tubes, plates, and bag with these numbers or your submission will be returned to you.
You should print out the PDF attachments from your acceptance email from the Core. You will need it for labeling, and you can then put it inside your submission bag.
You selected tubes or plates when you created your submission in MiCores. Detailed instructions for each are below.
Remember: Do not submit your entire sample. The AGC does not return unused aliquots.
1A) Preparing Tubes
- Aliquot your samples into 1.5 ml tubes.
- Securely close the tube caps.
Make sure that Core Sample Id numbers are clearly written on the top of each tube. Like this:
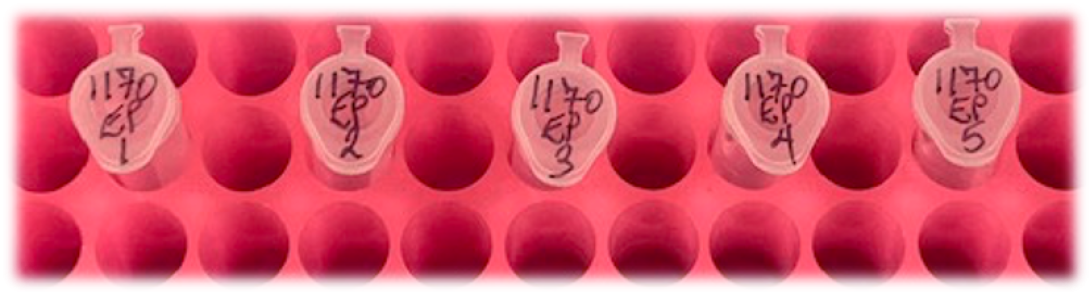
- See below for instructions on putting your samples into a plastic zip-top bag and getting them to the Core.
1B) Preparing Plates
- If submitting samples in plates, use 96-well, skirted PCR plates. Skirted plates help prevent bending, which can lead to leakage, during transit. The plates should be sealed with a foil cover capable of withstanding -80°C temperatures.
- Product Examples
- Plates:
- VWR #83007-374 VWR 96-Well PCR and Real Time PCR Plates; Half-skirt
- VWR #82006-704 VWR 96-Well PCR and Real Time PCR Plates; skirted
- Eppendorf #30129504 Eppendorf twin.tec PCR Plate 96 LoBind, semi-skirted, PCR clean
- Plates:
- Seals:
- VWR #30623-776 AlumaSeal 96 Sealing Foil (EXCEL), Nonsterile
- Corning #6570 Corning 96-well Microplate Aluminum Sealing Tape, Nonsterile
- Product Examples
- Aliquot your samples into plates using the plate map included in the acceptance email from the Core.
Note that your samples will be be arranged vertically (in columns) as shown here:

- Follow the instructions below on putting your samples into a plastic zip-top bag and getting them to the Core.
- Using a zip-top bag that you supply, write the Submission Id on the bag in permanent waterproof pen.
- Place all tubes or plates into the zip-top bag.
- Place your printed attachments inside the bag.

Do NOT place dry ice in the drop-off freezers. Samples submitted to the remote freezer locations (MSRBII C555 and NCRC Dock) are couriered to the AGC on dry ice M-F afternoons.
If the drop-off locations at MSRB II or NCRC014-122 are closed because of a weather incident, you can still drop off samples at NCRC Building 90 Dock (details below) daily between 7am and 4:30pm, as long as NCRC is open.
Sample drop-off is contactless. Drop-off instructions depend on the submission location:
If you work in the MSB complex, you can drop samples off in our MSRB II C555 location.
The MSRB II C555 (equipment corridor) location is accessible 24/7.

IMPORTANT: You must first aliquot your samples and bag them. See sample preparation instructions for more information.

- Bring the appropriately bagged aliquots to MSRB II, C555.
Put the bagged/boxed samples in the large “Advanced Genomics Core Sample Submission Freezer” in one of the metal baskets. Please note that you no longer need to enter info into a sample drop-off log.

- A courier transports samples from MSRB to the AGC on dry ice most weekdays at about 2:00 pm. Note that the exact date and time may change because of weather and other external factors.
When the core receives your bag, you will get an email notification from our system.
If you have questions, fill out our online Consultation Request Form.
If you work in U-M buildings other than the NCRC or MSRB complexes, or the AGC drop-off locations in NCRC Building 14 or MSRB are closed for a weather emergency, you can drop your samples at the NCRC dock (building 90).
NCRC Building 90 Dock is open workdays, 7:00am to 4:30pm.
Watch a video of how to find NCRC Building 90 dock.
IMPORTANT: You must first aliquot your samples and bag them. See sample preparation instructions for more information.

Bring the appropriately bagged/boxed samples to the NCRC dock located in building 90. Watch a video to see how to find NCRC 90 Dock.
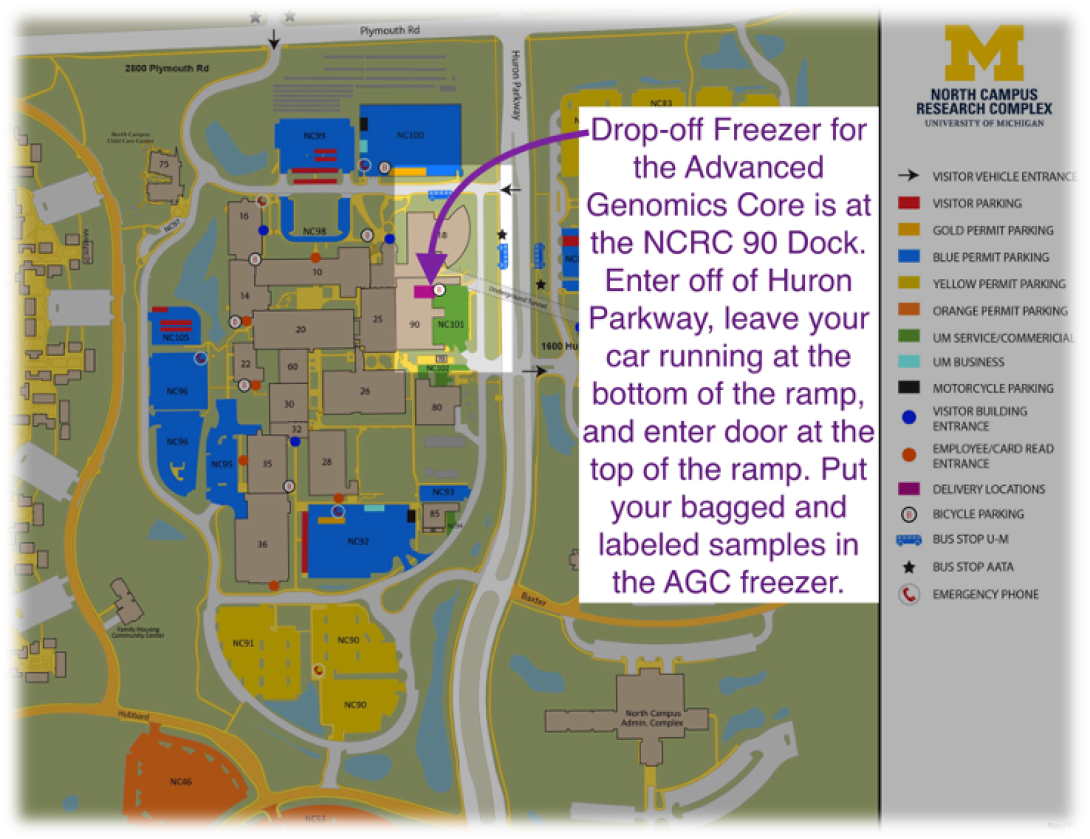

Enter via the B90 NE Exterior door leading to the dock GSS Security station.

Place the bagged/boxed samples in the freezer labeled AGC Sample Submission Freezer.

The drop off location will be accessible 7:00am – 4:30pm, Monday – Friday. Samples are transported on dry ice to the AGC daily.
If you have questions, fill out our online Consultation Request Form.
If you have access to NCRC Building 14, you can submit samples directly to the core.
IMPORTANT: You must first aliquot your samples and bag them. See sample preparation instructions for more information.

Bring samples and submission paperwork to the AGC drop-off window (NCRC Building 14, Room 122).

Following instructions on the freezer, place samples in the appropriate location.

When the core processes your bag, you will get an email notification from our system.

If you have questions, fill out our online Consultation Request Form.
If you do not have access to NCRC Building 14 or MSRB II
Customers who do not have access to NCRC or MSRB can either ship their samples to us, or drop them off at a freezer at the dock in NCRC Building 90.
- IMPORTANT: You must first aliquot your samples and bag them. See sample preparation instructions for more information.
- To drop off at the dock at NCRC Building 90 (follow the instructions above). We also created a video to help you find the dock at building 90.
- To ship your samples to us, send them to:
U-M Advanced Genomics Core
NCRC Building 14 Rm 122
University of Michigan
2800 Plymouth Rd
Ann Arbor, MI 48109-2800
Phone: 734-764-8068
| Assay | Buffer | Amount |
| 10X Genomics 3', 5' or Immune Profiling | Most standard medias with no more than 2% BSA, 10% FBS, and no EDTA | 50K cells in 40-50ul buffer |
| 10X Genomics ATAC or Multiome (GEX+ATAC) | 10X Genomics Nuclei Buffer* | 50K nuclei in 10-20ul buffer |
| 10X Genomics Single Cell RNA Flex | 10X Genomics Quenching/Resuspension Buffer* | 50K cells or 5-50mg tissue |
| ParseBiosciences EverCode | Parse Biosciences fixation solution* | 100K cells |
| Direct Cell Lysis | Vendor provided buffer in 96-well plate* | Full 96-well plate |
*These buffers can be picked up/purchased from AGC
| Assay | Slide Type | Thickness |
| 10X Genomics CytAssist | Charged glass slide | 5um FFPE; 10um Frozen |
| Nanostring GeoMx DSP | Charged glass slide | 5um FFPE; 10um Frozen |
| Curio Biosciences CurioSeeker | CurioSeeker Tile* | 10um Fresh Frozen |
| 10X Genomics Xenium | Xenium slide* | 5um FFPE; 10um Frozen |
*These can picked picked up/purchased from or sectioned by AGC
Make your appointment via the AGC MiCores site – “Schedule Equipment” -> “Single Cell Appointment Request”
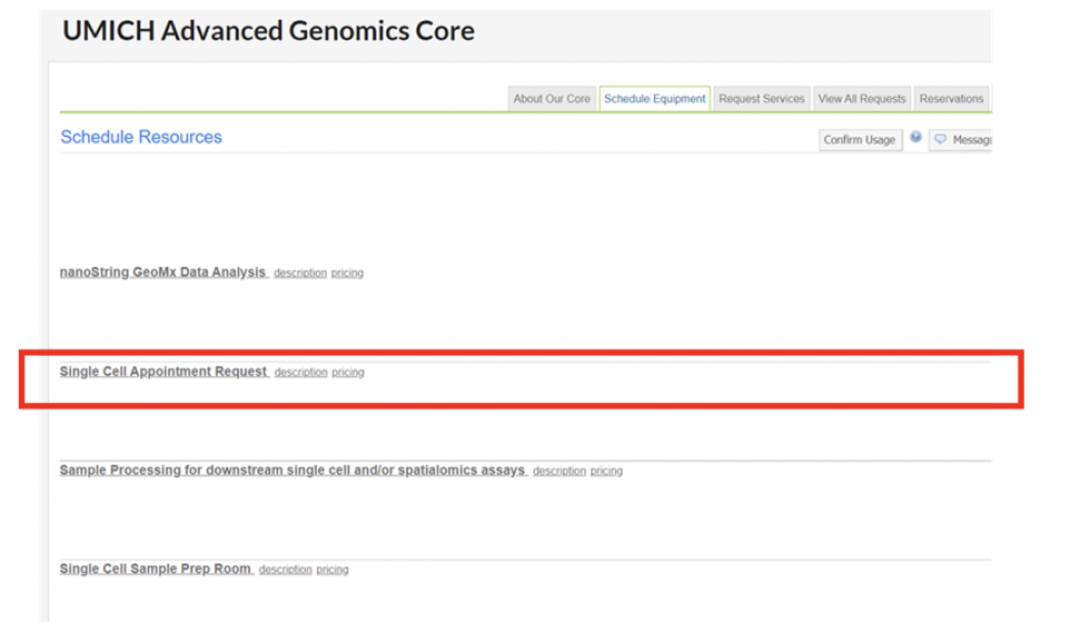
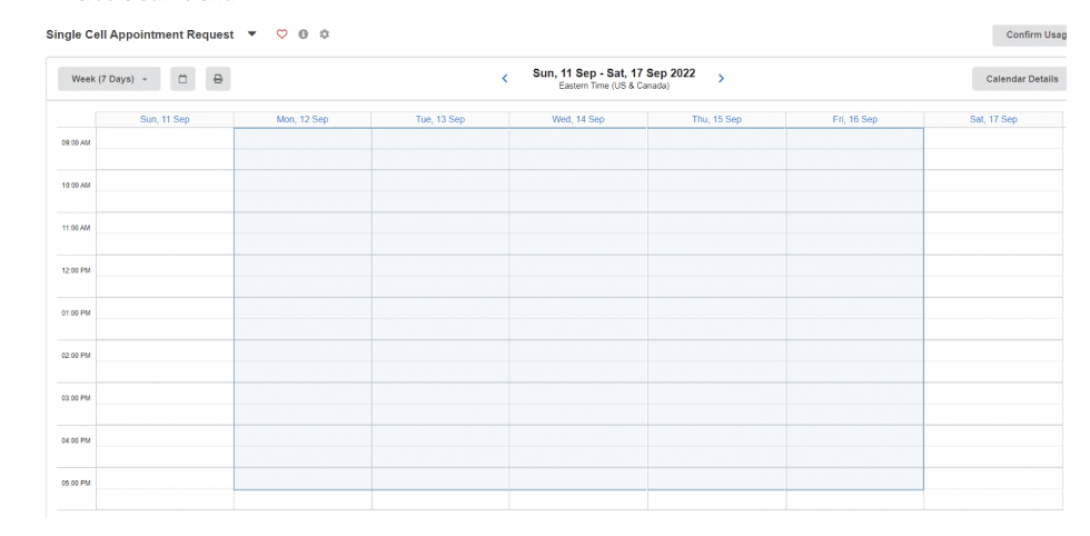
You may select any time within the blue boxes. If you have fewer than 6 samples, a 30 minute time slot is sufficient.
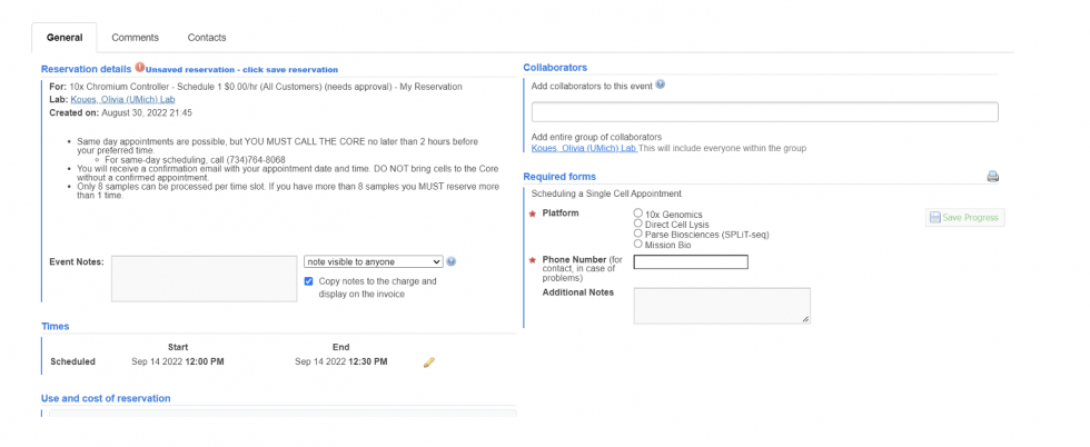
Fill out the form. We have a large number of options, so please look them over carefully. Viability/Count checks can be found under the “10X Genomics” options. Please note the “Comments” tab at the top – if you need to cancel your appointment after it has been confirmed, or anything else comes up, you can use this tab to communicate with us.
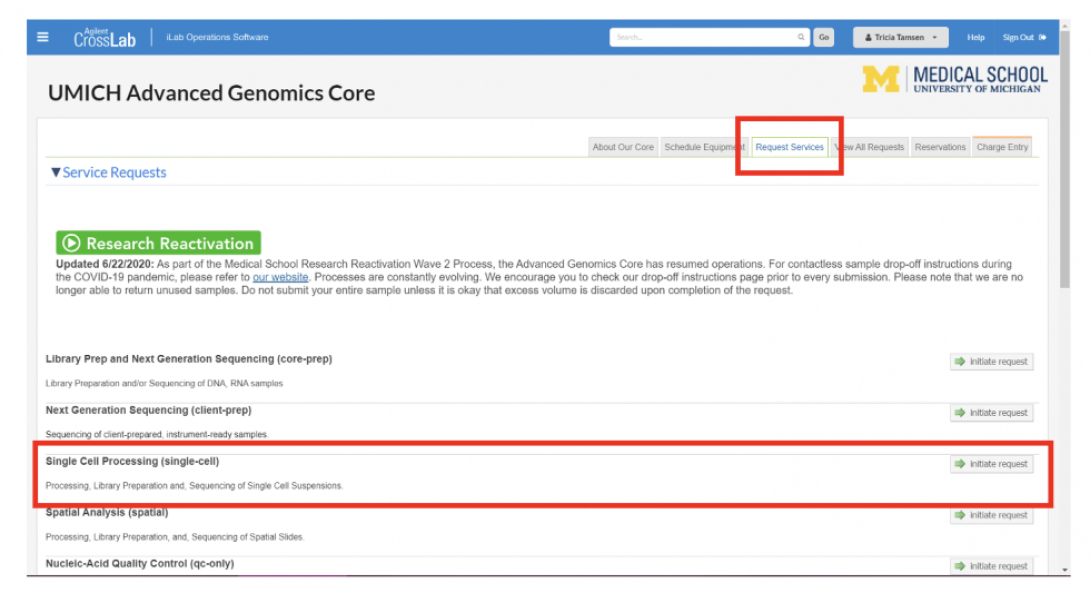
Submit your service request on the AGC MiCores site – “Request Services” -> “Single Cell Processing.” This MUST be done before you bring your cells to the Core. Most people fill out their service request 2-24 hours before their appointment time. If you have not created a service request before bringing your cells, we cannot accept them.
The request form will ask you for your analysis type, read depth (sequencing depth), targeted number of cells (1,000-10,000), and your sample names. When that has been submitted, you will receive an email with the numbers that correspond to each sample. Write your sample numbers on the top of your sample tubes. If your sample numbers are not on the tops of your tubes, we cannot accept them.

Package your samples in either a plastic bag or 50ml conical tube, then inside a cooler on ice. If your samples are not in an appropriate secondary container, we cannot accept them. This protects both you and our technicians, and is mandated by EHS. Pictures of two acceptable submissions are below.
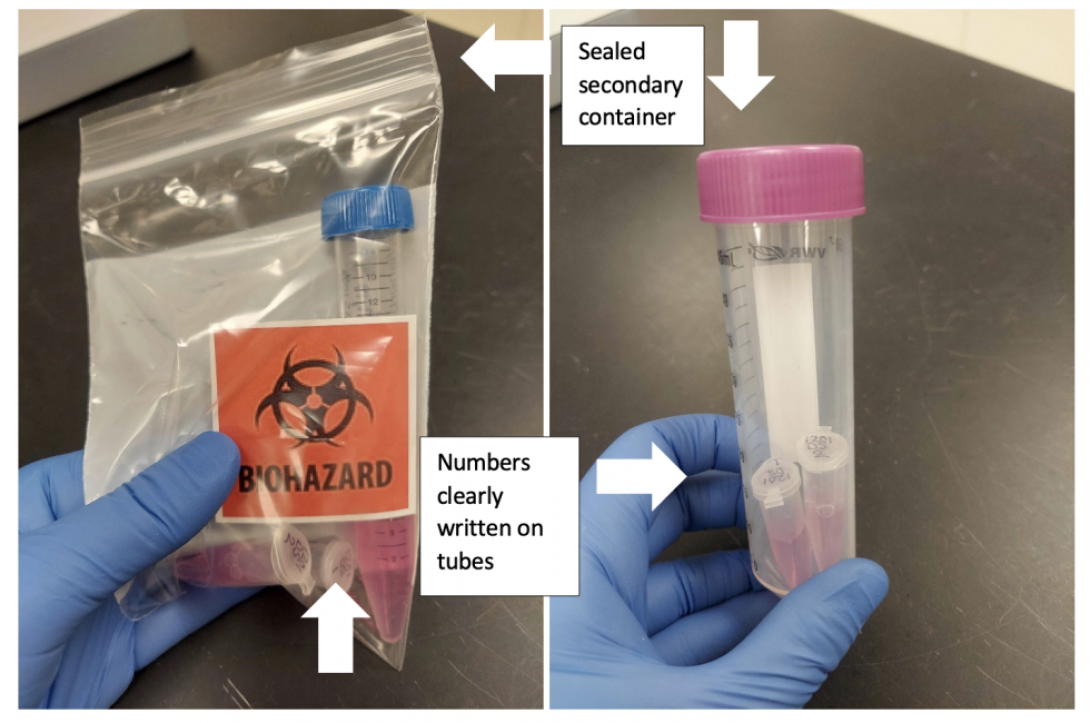
Come to NCRC. Guest services is now staffing the lobbies at both Building 18 and Building 32. Show your MCard to gain access. You will then need to make your way to the lab in Building 14. Note that the most direct path through the complex is to enter through Building 32.
Ring the Single Cell and Spatial doorbell when you arrive. We will take your samples and check the concentration and viability while you wait. We will discuss the results with you so that you can make any decisions to proceed immediately.
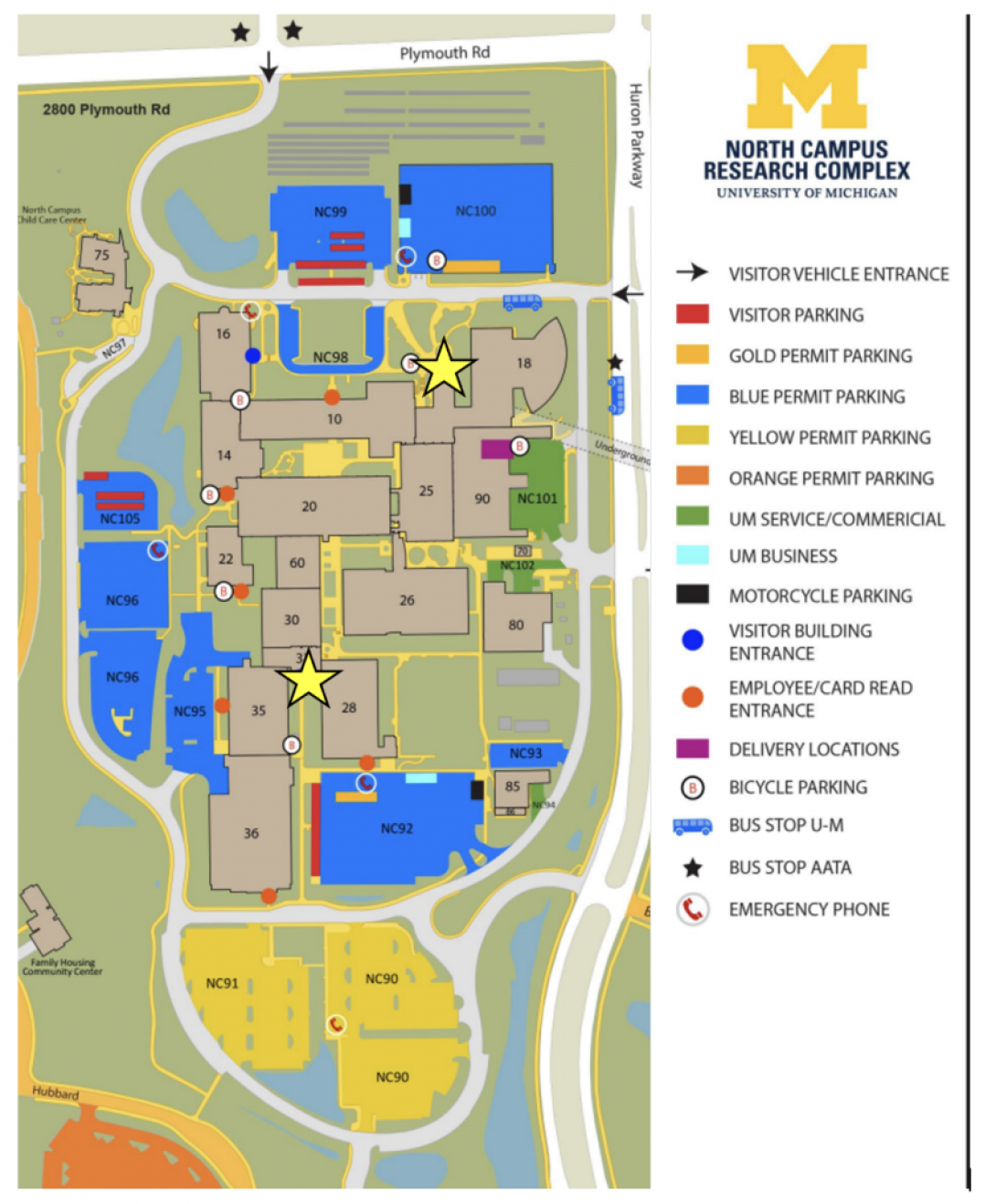
Walk up the extremely long walkway to the door to Building 32. Head to the right and show your MCard to the Guest Services agent. Through the lobby, turn left down the long hallway. You will pass the 24/7 Market and walk up one small set of stairs.

Walk straight until you reach the courtyard. Turn left. Walk straight until you have entered Building 14. Once you see the recycling gondola, look to your right. AGC Single Cell and Spatial drop-off is at Room 171. Ring the doorbell and someone will be with you shortly.

- Appointments are required to submit single cell or spatial samples. Reservations are made via MiCores.
- Submission: Samples should be placed in a secondary container, e.g. a small plastic biohazard bag, for transport to the AGC.
University of Michigan
2800 Plymouth Rd.
Ann Arbor, MI 48109-2800